Portfolio
BIG-TB: A Benchmark Dataset for Genomic Resistance Prediction and Interpretability
A multimodal benchmark of 17,000 Mycobacterium tuberculosis isolates for antibiotic resistance modeling and causal variant discovery.
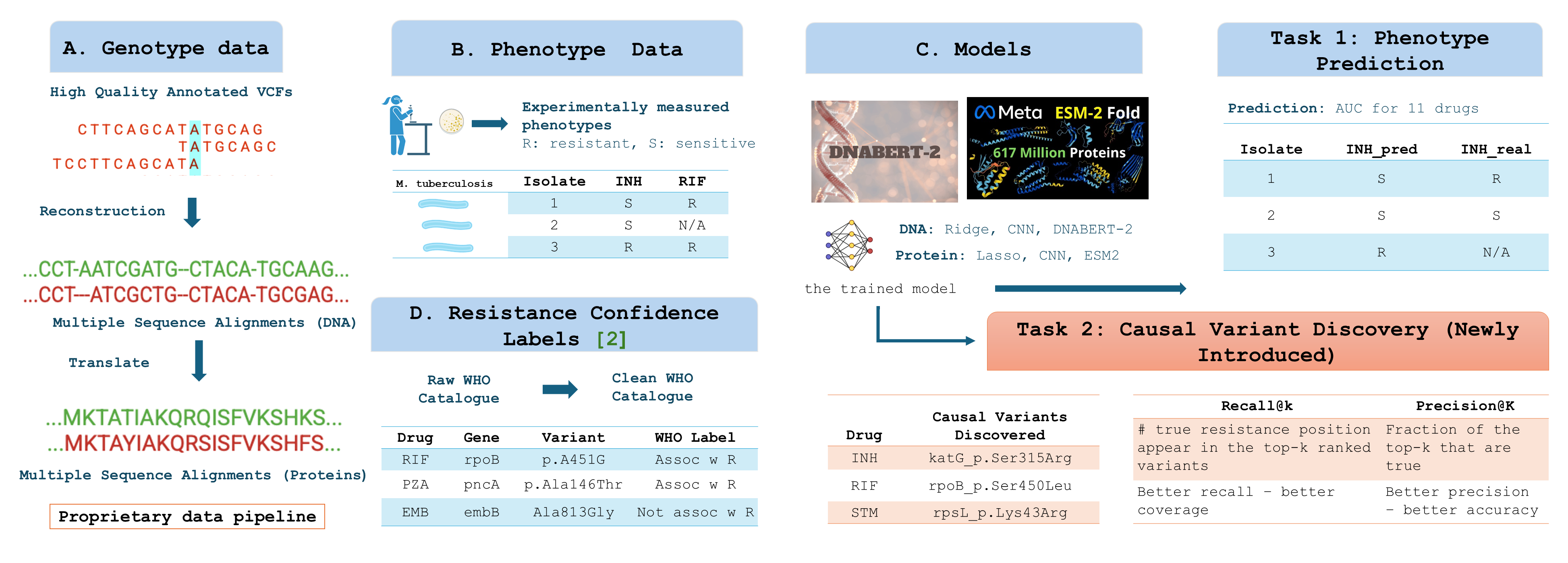
BIG-TB is a large-scale, multimodal benchmark dataset designed to advance machine-learning-based antibiotic resistance prediction and biological interpretability in Mycobacterium tuberculosis.
The dataset integrates genomic, protein, and structural features across 11 WHO-priority drugs and enables rigorous evaluation of both predictive accuracy and causal variant recovery.
🔬 Key Contributions
- Curated 17K isolates with unified resistance labels and high-confidence mutation annotations from WHO 2023 and 2021 catalogues.
- Developed reproducible pipelines for sequence, structure, and evolutionary data integration (DNA + protein + 3D features).
- Introduced interpretability benchmarks via Precision@k and SHAP-based variant recovery.
- Released accompanying code and documentation to promote transparency and reproducibility.
🧠 Methods
Models include:
- CNNs and Transformers for sequence-level modeling
- ESM-based protein embeddings and PCA-compressed representations
- Linear fused-ridge baselines for interpretable structure-aware learning
🏆 Impact
Presented as a spotlight talk at the Machine Learning for Computational Biology (MLCB) Workshop 2025, co-located with NeurIPS.
BIG-TB serves as a foundation for evaluating biological faithfulness in ML systems and fostering explainable AI for global health.